Wod bgc ragged example
Wod bgc ragged example example.
4 minute read
An example of using COAsT to analysis observational profile data alongside gridded NEMO data.
Load modules
import coast
import glob # For getting file paths
import gsw
import matplotlib.pyplot as plt
import datetime
import numpy as np
import xarray as xr
import coast._utils.general_utils as general_utils
import scipy as sp
# ====================== UNIV PARAMS ===========================
path_examples = "./example_files/"
path_config = "./config/"
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/pydap/lib.py:5: DeprecationWarning: pkg_resources is deprecated as an API. See https://setuptools.pypa.io/en/latest/pkg_resources.html
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/pkg_resources/__init__.py:2871: DeprecationWarning: Deprecated call to `pkg_resources.declare_namespace('pydap')`.
Implementing implicit namespace packages (as specified in PEP 420) is preferred to `pkg_resources.declare_namespace`. See https://setuptools.pypa.io/en/latest/references/keywords.html#keyword-namespace-packages
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/pkg_resources/__init__.py:2871: DeprecationWarning: Deprecated call to `pkg_resources.declare_namespace('pydap.responses')`.
Implementing implicit namespace packages (as specified in PEP 420) is preferred to `pkg_resources.declare_namespace`. See https://setuptools.pypa.io/en/latest/references/keywords.html#keyword-namespace-packages
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/pkg_resources/__init__.py:2350: DeprecationWarning: Deprecated call to `pkg_resources.declare_namespace('pydap')`.
Implementing implicit namespace packages (as specified in PEP 420) is preferred to `pkg_resources.declare_namespace`. See https://setuptools.pypa.io/en/latest/references/keywords.html#keyword-namespace-packages
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/pkg_resources/__init__.py:2871: DeprecationWarning: Deprecated call to `pkg_resources.declare_namespace('pydap.handlers')`.
Implementing implicit namespace packages (as specified in PEP 420) is preferred to `pkg_resources.declare_namespace`. See https://setuptools.pypa.io/en/latest/references/keywords.html#keyword-namespace-packages
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/pkg_resources/__init__.py:2350: DeprecationWarning: Deprecated call to `pkg_resources.declare_namespace('pydap')`.
Implementing implicit namespace packages (as specified in PEP 420) is preferred to `pkg_resources.declare_namespace`. See https://setuptools.pypa.io/en/latest/references/keywords.html#keyword-namespace-packages
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/pkg_resources/__init__.py:2871: DeprecationWarning: Deprecated call to `pkg_resources.declare_namespace('pydap.tests')`.
Implementing implicit namespace packages (as specified in PEP 420) is preferred to `pkg_resources.declare_namespace`. See https://setuptools.pypa.io/en/latest/references/keywords.html#keyword-namespace-packages
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/pkg_resources/__init__.py:2350: DeprecationWarning: Deprecated call to `pkg_resources.declare_namespace('pydap')`.
Implementing implicit namespace packages (as specified in PEP 420) is preferred to `pkg_resources.declare_namespace`. See https://setuptools.pypa.io/en/latest/references/keywords.html#keyword-namespace-packages
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/pkg_resources/__init__.py:2871: DeprecationWarning: Deprecated call to `pkg_resources.declare_namespace('sphinxcontrib')`.
Implementing implicit namespace packages (as specified in PEP 420) is preferred to `pkg_resources.declare_namespace`. See https://setuptools.pypa.io/en/latest/references/keywords.html#keyword-namespace-packages
load and preprocess profile and model data
fn_wod_var = path_examples + "WOD_example_ragged_standard_level.nc"
fn_wod_config = path_config + "example_wod_profiles.json"
wod_profile_1d = coast.Profile(config=fn_wod_config)
wod_profile_1d.read_wod(fn_wod_var)
./config/example_wod_profiles.json
Reshape into 2D. Choose which observed variables you want.
var_user_want = ["salinity", "temperature", "nitrate", "oxygen", "dic", "phosphate", "alkalinity"]
wod_profile = coast.Profile.reshape_2d(wod_profile_1d, var_user_want)
Depth OK reshape successful
salinity
observed variable exist
OK reshape successful
temperature
observed variable exist
OK reshape successful
nitrate
variable not in observations
oxygen
observed variable exist
OK reshape successful
dic
observed variable exist
OK reshape successful
phosphate
observed variable exist
OK reshape successful
alkalinity
variable not in observations
Keep subset.
wod_profile_sub = wod_profile.subset_indices_lonlat_box(lonbounds=[90, 120], latbounds=[-5, 5])
#wod_profile_sub.dataset # uncomment to print data object summary
SEAsia read BGC. Note in this simple test nemo data are only for 3 months from 1990 so the comparisons are not going to be correct but just as a demo.
fn_seasia_domain = path_examples + "coast_example_domain_SEAsia.nc"
fn_seasia_config_bgc = path_config + "example_nemo_bgc.json"
fn_seasia_var = path_examples + "coast_example_SEAsia_BGC_1990.nc"
seasia_bgc = coast.Gridded(
fn_data=fn_seasia_var, fn_domain=fn_seasia_domain, config=fn_seasia_config_bgc, multiple=True
)
Domain file does not have mask so this is just a trick.
seasia_bgc.dataset["landmask"] = seasia_bgc.dataset.bottom_level == 0
seasia_bgc.dataset = seasia_bgc.dataset.rename({"depth_0": "depth"})
model_profiles = wod_profile_sub.obs_operator(seasia_bgc)
#model_profiles.dataset # uncomment to print data object summary
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/xarray/core/utils.py:494: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/xarray/core/utils.py:494: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/coast/data/profile.py:456: UserWarning: Converting non-nanosecond precision timedelta values to nanosecond precision. This behavior can eventually be relaxed in xarray, as it is an artifact from pandas which is now beginning to support non-nanosecond precision values. This warning is caused by passing non-nanosecond np.datetime64 or np.timedelta64 values to the DataArray or Variable constructor; it can be silenced by converting the values to nanosecond precision ahead of time.
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/coast/data/profile.py:456: UserWarning: Converting non-nanosecond precision timedelta values to nanosecond precision. This behavior can eventually be relaxed in xarray, as it is an artifact from pandas which is now beginning to support non-nanosecond precision values. This warning is caused by passing non-nanosecond np.datetime64 or np.timedelta64 values to the DataArray or Variable constructor; it can be silenced by converting the values to nanosecond precision ahead of time.
Remove any points that are far from model.
too_far = 5
keep_indices = model_profiles.dataset.interp_dist <= too_far
model_profiles = model_profiles.isel(id_dim=keep_indices)
wod_profile = wod_profile_sub.isel(id_dim=keep_indices)
#wod_profile.dataset # uncomment to print data object summary
Plot profiles
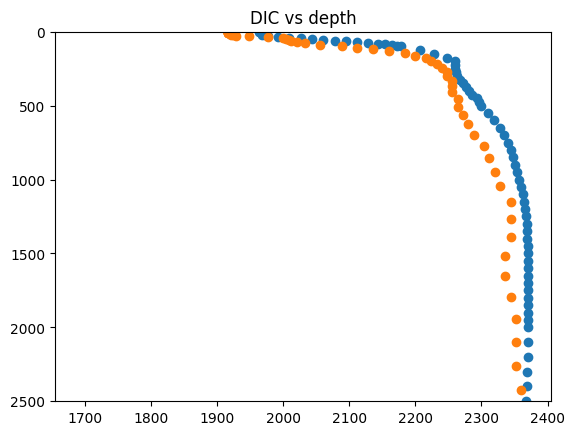
Transform observed DIC from mmol/l to mmol C/ m^3 that the model has.
fig = plt.figure()
plt.plot(1000 * wod_profile.dataset.dic[8, :], wod_profile.dataset.depth[8, :], linestyle="", marker="o")
plt.plot(model_profiles.dataset.dic[8, :], model_profiles.dataset.depth[:, 8], linestyle="", marker="o")
plt.ylim([2500, 0])
plt.title("DIC vs depth")
plt.show()
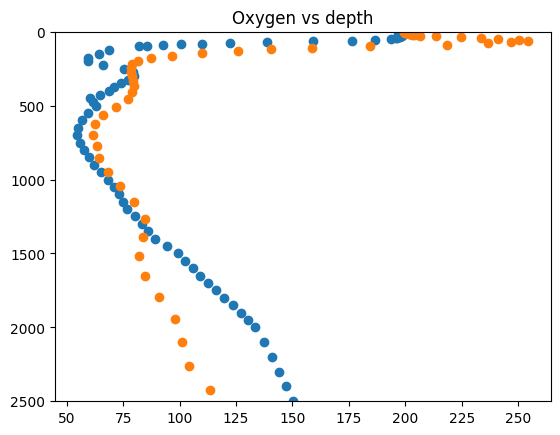
fig = plt.figure()
plt.plot(wod_profile.dataset.oxygen[8, :], wod_profile.dataset.depth[8, :], linestyle="", marker="o")
plt.plot(model_profiles.dataset.oxygen[8, :], model_profiles.dataset.depth[:, 8], linestyle="", marker="o")
plt.ylim([2500, 0])
plt.title("Oxygen vs depth")
plt.show()
Perform profile analysis to evaluate differences
Interpolate seasia to profile depths, using ProfileAnalysis class.
reference_depths = wod_profile.dataset.depth[20, :].values
model_profiles.dataset = model_profiles.dataset[["dic"]] / 1000
pa = coast.ProfileAnalysis()
model_interpolated = pa.interpolate_vertical(model_profiles, wod_profile)
Calculate differences.
differences = pa.difference(model_interpolated, wod_profile)
#differences.dataset.load() # uncomment to print data object summary
Feedback
Was this page helpful?
Glad to hear it!
Sorry to hear that. Please tell us how we can improve.
Last modified December 11, 2023: Updated Profile notebook pages from coast repo. (5e75920)