Seasonal decomp example
10 minute read
Overview
A function within the Process_data class that will decompose time series into trend, seasonal and residual components. The function is a wrapper that adds functionality to the seasonal_decompose function contained in the statsmodels package to make it more convenient for large geospatial datasets.
Specifically:
- Multiple time series spread across multiple dimensions, e.g. a gridded dataset, can be processed. The user simply passes in an xarray DataArray that has a “t_dim” dimension and 1 or more additional dimensions, for example gridded spatial dimensions
- Masked locations, such as land points, are handled
- A dask wrapper is applied to the function that a) supports lazy evaluation b) allows the dataset to be easily seperated into chunks so that processing can be carried out in parallel (rather than processing every time series sequentially)
- The decomposed time series are returned as xarray DataArrays within a single coast.Gridded object
An example
Below is an example using the coast.Process_data.seasonal_decomposition function with the example data. Note that we will artifically extend the length of the example data time series for demonstrative purposes.
Begin by importing coast, defining paths to the data, and loading the example data into a gridded object:
import coast
import numpy as np
import xarray as xr
# Path to a data file
root = "./"
dn_files = root + "./example_files/"
fn_nemo_dat = dn_files + "coast_example_nemo_data.nc"
# Set path for domain file if required.
fn_nemo_dom = dn_files + "coast_example_nemo_domain.nc"
# Set path for model configuration file
config = root + "./config/example_nemo_grid_t.json"
# Read in data (This example uses NEMO data.)
grd = coast.Gridded(fn_nemo_dat, fn_nemo_dom, config=config)
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/pydap/lib.py:5: DeprecationWarning: pkg_resources is deprecated as an API. See https://setuptools.pypa.io/en/latest/pkg_resources.html
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/pkg_resources/__init__.py:2871: DeprecationWarning: Deprecated call to `pkg_resources.declare_namespace('pydap')`.
Implementing implicit namespace packages (as specified in PEP 420) is preferred to `pkg_resources.declare_namespace`. See https://setuptools.pypa.io/en/latest/references/keywords.html#keyword-namespace-packages
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/pkg_resources/__init__.py:2871: DeprecationWarning: Deprecated call to `pkg_resources.declare_namespace('pydap.responses')`.
Implementing implicit namespace packages (as specified in PEP 420) is preferred to `pkg_resources.declare_namespace`. See https://setuptools.pypa.io/en/latest/references/keywords.html#keyword-namespace-packages
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/pkg_resources/__init__.py:2350: DeprecationWarning: Deprecated call to `pkg_resources.declare_namespace('pydap')`.
Implementing implicit namespace packages (as specified in PEP 420) is preferred to `pkg_resources.declare_namespace`. See https://setuptools.pypa.io/en/latest/references/keywords.html#keyword-namespace-packages
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/pkg_resources/__init__.py:2871: DeprecationWarning: Deprecated call to `pkg_resources.declare_namespace('pydap.handlers')`.
Implementing implicit namespace packages (as specified in PEP 420) is preferred to `pkg_resources.declare_namespace`. See https://setuptools.pypa.io/en/latest/references/keywords.html#keyword-namespace-packages
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/pkg_resources/__init__.py:2350: DeprecationWarning: Deprecated call to `pkg_resources.declare_namespace('pydap')`.
Implementing implicit namespace packages (as specified in PEP 420) is preferred to `pkg_resources.declare_namespace`. See https://setuptools.pypa.io/en/latest/references/keywords.html#keyword-namespace-packages
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/pkg_resources/__init__.py:2871: DeprecationWarning: Deprecated call to `pkg_resources.declare_namespace('pydap.tests')`.
Implementing implicit namespace packages (as specified in PEP 420) is preferred to `pkg_resources.declare_namespace`. See https://setuptools.pypa.io/en/latest/references/keywords.html#keyword-namespace-packages
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/pkg_resources/__init__.py:2350: DeprecationWarning: Deprecated call to `pkg_resources.declare_namespace('pydap')`.
Implementing implicit namespace packages (as specified in PEP 420) is preferred to `pkg_resources.declare_namespace`. See https://setuptools.pypa.io/en/latest/references/keywords.html#keyword-namespace-packages
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/pkg_resources/__init__.py:2871: DeprecationWarning: Deprecated call to `pkg_resources.declare_namespace('sphinxcontrib')`.
Implementing implicit namespace packages (as specified in PEP 420) is preferred to `pkg_resources.declare_namespace`. See https://setuptools.pypa.io/en/latest/references/keywords.html#keyword-namespace-packages
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/xarray/core/dataset.py:282: UserWarning: The specified chunks separate the stored chunks along dimension "time_counter" starting at index 2. This could degrade performance. Instead, consider rechunking after loading.
The loaded example data only has 7 time stamps, the code below creates a new (fake) extended temperature variable with 48 monthly records. This code is not required to use the function, it is only included here to make a set of time series that are long enough to be interesting.
# create a 4 yr monthly time coordinate array
time_array = np.arange(
np.datetime64("2010-01-01"), np.datetime64("2014-01-01"), np.timedelta64(1, "M"), dtype="datetime64[M]"
).astype("datetime64[s]")
# create 4 years of monthly temperature data based on the loaded data
temperature_array = (
(np.arange(0, 48) * 0.05)[:, np.newaxis, np.newaxis, np.newaxis]
+ np.random.normal(0, 0.1, 48)[:, np.newaxis, np.newaxis, np.newaxis]
+ np.tile(grd.dataset.temperature[:-1, :2, :, :], (8, 1, 1, 1))
)
# create a new temperature DataArray
temperature = xr.DataArray(
temperature_array,
coords={
"t_dim": time_array,
"depth_0": grd.dataset.depth_0[:2, :, :],
"longitude": grd.dataset.longitude,
"latitude": grd.dataset.latitude,
},
dims=["t_dim", "z_dim", "y_dim", "x_dim"],
)
/tmp/ipykernel_3937/2929135463.py:14: UserWarning: Converting non-nanosecond precision datetime values to nanosecond precision. This behavior can eventually be relaxed in xarray, as it is an artifact from pandas which is now beginning to support non-nanosecond precision values. This warning is caused by passing non-nanosecond np.datetime64 or np.timedelta64 values to the DataArray or Variable constructor; it can be silenced by converting the values to nanosecond precision ahead of time.
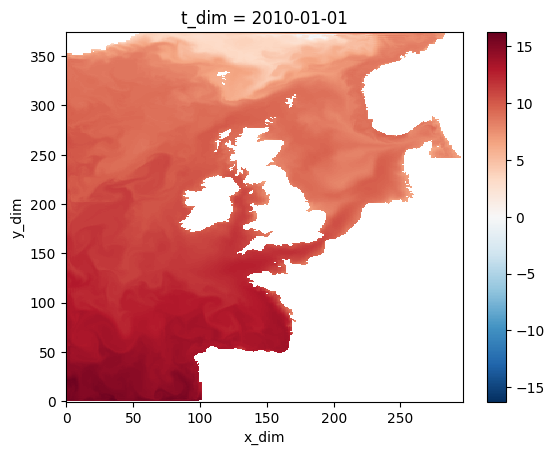
Check out the new data
#temperature # uncomment to print data object summary
temperature[0,0,:,:].plot()
<matplotlib.collections.QuadMesh at 0x7fc9f43a0f70>
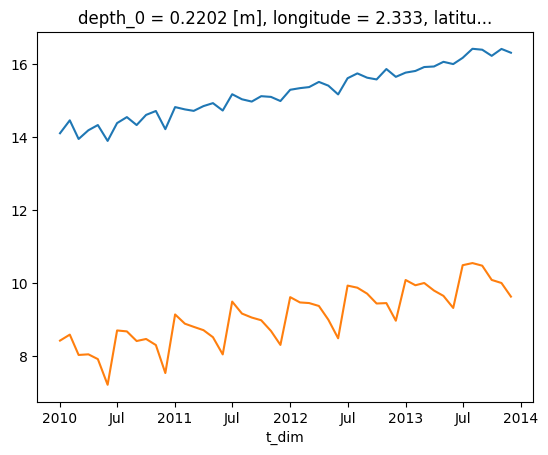
Check out time series at 2 different grid points
temperature[:,0,50,50].plot()
temperature[:,0,200,200].plot()
[<matplotlib.lines.Line2D at 0x7fc9f0f97f40>]
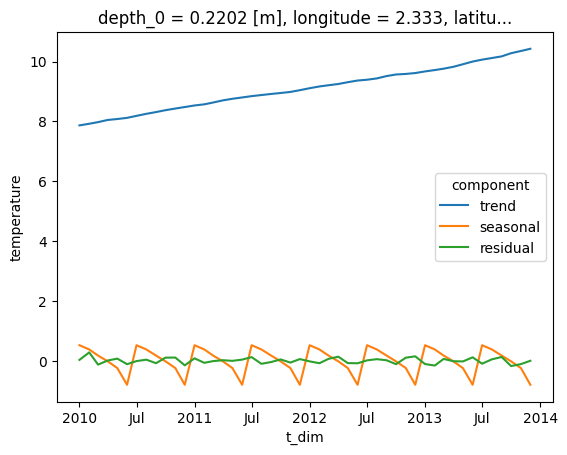
Create a coast.Process_data object, and call the seasonal_decomposition function, passing in the required arguments. The first two arguments are:
- The input data, here the temperature data as an xarray DataArray
- The number of chuncks to split the data into. Here we split the data into 2 chunks so that the dask scheduler will try to run 4 processes in parallel
The remaining arguments are keyword arguments for the underlying statsmodels.tsa.seasonal.seasonal_decompose function, which are documented on the statsmodels documentation pages. Here we specify:
- three The type of model, i.e. an additive model
- The period of the seasonal cycle, here it is 6 months
- Extrapolate the trend component to cover the entire range of the time series (this is required because the trend is calculated using a convolution filter)
proc_data = coast.Process_data()
grd = proc_data.seasonal_decomposition(temperature, 2, model="additive", period=6, extrapolate_trend="freq")
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/coast/_utils/logging_util.py:21: DeprecationWarning: ProcessData is deprecated: Please use the 'ProcessData' class instead. This name will change in the next release.
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/coast/_utils/process_data.py:114: DeprecationWarning: Supplying chunks as dimension-order tuples is deprecated. It will raise an error in the future. Instead use a dict with dimension names as keys.
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/coast/_utils/process_data.py:116: DeprecationWarning: Supplying chunks as dimension-order tuples is deprecated. It will raise an error in the future. Instead use a dict with dimension names as keys.
/usr/share/miniconda/envs/coast/lib/python3.10/site-packages/coast/_utils/process_data.py:118: DeprecationWarning: Supplying chunks as dimension-order tuples is deprecated. It will raise an error in the future. Instead use a dict with dimension names as keys.
The returned xarray Dataset contains the decomposed time series (trend, seasonal, residual) as dask arrays
#grd.dataset # uncomment to print data object summary
Execute the computation
grd.dataset.compute()
<xarray.Dataset> Dimensions: (t_dim: 48, z_dim: 2, y_dim: 375, x_dim: 297) Coordinates:
-
t_dim (t_dim) datetime64[ns] 2010-01-01 2010-02-01 … 2013-12-01 depth_0 (z_dim, y_dim, x_dim) float32 0.5 0.5 0.5 0.5 … 1.5 1.5 1.5 1.5 longitude (y_dim, x_dim) float32 -19.89 -19.78 -19.67 … 12.78 12.89 13.0 latitude (y_dim, x_dim) float32 40.07 40.07 40.07 40.07 … 65.0 65.0 65.0 Dimensions without coordinates: z_dim, y_dim, x_dim Data variables: trend (t_dim, z_dim, y_dim, x_dim) float64 nan nan nan … nan nan nan seasonal (t_dim, z_dim, y_dim, x_dim) float64 nan nan nan … nan nan nan residual (t_dim, z_dim, y_dim, x_dim) float64 nan nan nan … nan nan nan